Cows can adapt themselves to a fluctuating sodium content in their feed. How they do that was so far a secret. Researchers from Goethe University have now discovered a bacterium in the microbiome of the rumen which has a new type of cell respiration.
The cow can only process grass in its rumen with the help of billions of microorganisms. An entire zoo of bacteria, archaea and protozoa works there like on a production line: First of all, these single-cell organisms break down the cellulose, a polysaccharide. Other bacteria ferment the sugars released into fatty acids, alcohols and gases, such as hydrogen and carbon dioxide. Finally, methanogenic archaea transform these two gases into methane.
An average cow produces about 110 liters of methane per day. It escapes from its mouth through rumination, but also mixes again with partly digested food. As a result, the sodium content of the grass pulp can fluctuate to a considerable degree (between 60 and 800 millimoles of sodium chloride (NaCl) per liter).
A German-American research team has now discovered how the ruminal bacteria adapt to these extreme fluctuations in sodium content: “Bioinformatic analyses of the genome of ruminal bacteria led our American colleague Tim Hackmann to assume that some ruminal bacteria have two different respiratory circuits. One of them functions with sodium ions and the other without,” explains Professor Volker Müller from the Department of Molecular Microbiology and Bioenergetics at Goethe University. That is why Müller suggested to his doctoral researcher Marie Schölmerich that she study a typical representative in the microbiome of ruminants: the bacterium Pseudobutyrivibrio ruminis.
Together with undergraduate student Judith Dönig and Master’s student Alexander Katsyv, Marie Schölmerich cultivated the bacterium. Indeed, they were able to corroborate both respiratory circuits. As the researchers report in the current issue of the Proceedings of the National Academy of Sciences (PNAS), the electron carrier ferredoxin (Fd) is reduced during sugar oxidation. Reduced ferredoxin drives both respiratory circuits.
The one respiratory circuit comprises the enzyme complex Fd:NAD+ oxidoreductase (Rnf complex). It uses energy to transport sodium ions out of the cell. When they re-enter the cell, the sodium ions trigger an ATP synthase, so that ATP is produced. This respiratory circuit only works in the presence of sodium ions.
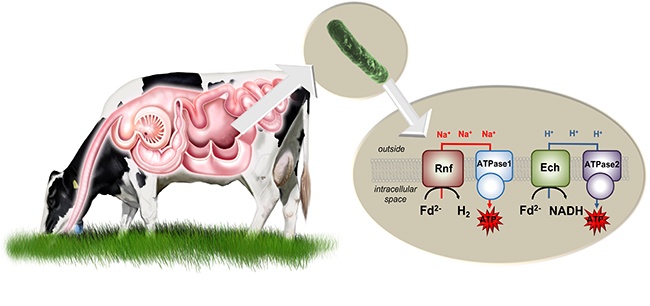
In the absence of sodium ions, the bacterium forms an alternative respiratory circuit with another enzyme complex: The Ech hydrogenase (synonymous: Fd:H+ oxidoreductase) produces hydrogen and pumps protons out of the cell. If these re-enter the cell via a second ATP synthase that accepts protons but not sodium ions, ATP is also produced.
“This is the first bacterium so far in which these two simple, completely different respiratory circuits have been corroborated, but our bioinformatic analyses suggest that they are also found in other bacteria,” explains Marie Schölmerich. “It seems, therefore, that this adaptation strategy is more widespread,” she assumes.
Interestingly, both enzyme complexes (Rnf and Ech) were also discovered in bacteria which are old in terms of evolutionary biology. Professor Müller’s research group has examined them in depth, but always only found one of the two enzyme complexes and never both together. “We’re now going to use synthetic microbiology methods to produce hybrids of bacteria that contain both complexes in order to optimize them for biotechnological processes. In this way, we can raise the cellular ATP content, which will make it possible to produce products of a higher quality,” explains Professor Müller. The intention is to use the respiratory circuits to recover valuable substances through the fermentation of synthesis gas. This is the subject of the trials being conducted in the framework of a project sponsored by the Federal Ministry of Education and Research
Publication: Schölmerich, M.C., Katsyv, A., Dönig, J., Hackmann, T., Müller, V. (20XX). Energy conservation involving two respiratory circuits. Proc. Natl. Acad. Sci. U.S.A. https://doi.org/10.1073/pnas.1914939117







