Volker Zickermann and Eric Helfrich are part of the excellence cluster initiative SCALE (Subcellular Architecture of Life). Whereas the former will contribute his expertise researching a protein complex in mitochondria, the cell’s power plants, the latter possesses specialist knowledge in the search for as yet unknown natural substances that could lead to new antibiotics.
Biology also has dark matter
Bacteria, fungi and plants produce certain substances in order to communicate with one another or to protect themselves from predators. These natural substances are indispensable to medicine – after all, more than 50 percent of all licensed medicines are either based on or were inspired by them. Bacteria are especially important producers of natural substances; in fact, most antibiotics developed to combat bacterial infections originated from bacteria. There is, however, a huge problem, as resistance to antibiotics is quickly becoming ever more widespread. “If we don’t counteract this development, in the future many people will die from bacterial infections because our antibiotics will be ineffective against multidrug-resistant pathogens,” says Eric Helfrich, head of the Natural Product Genomics department at the LOEWE Centre for translational biodiversity genomics [editor’s note: LOEWE is the research promotion program that has been used by the federal state of Hessen since 2008 to set research policy trends, the objective being to give a sustainable boost to Hesse’s research landscape. LOEWE stands for “Landes-Offensive zur Entwicklung wissenschaftlich-ökonomischer Exzellenz” (state program for the development of scientific and economic excellence)]. Helfrich hopes his team will help avert this horror scenario by using a computer-aided approach called genome mining to discover previously unknown natural bacterial products. Helfrich’s group develops machine-learning algorithms that analyze bacterial genome sequences with a view towards determining their potential for the production of natural substances. These products could become the lead structure for developing future medicines to combat infectious diseases.
Mining the bacterial genome is relatively easy, Helfrich explains. “There are certain genes coding for enzymes that build and modify the natural substance’s basic framework. In plants these genes are scattered across the entire genome like a mosaic – which makes it more difficult to identify them. In bacteria, by contrast, they have adjacent positions and form biosynthesis gene clusters. These are the clusters were looking for – or, to be precise, the ones that were overlooked by previous algorithms.” The Helfrich team starts by developing hypotheses about which types of gene clusters may have “fallen through the cracks” before, and then trains the algorithms to identify them. “Doing so makes the biosynthetic dark matter accessible, that is, the hitherto undiscovered clusters that serve as construction plans for building natural products.”
One difficulty in the search for new natural substances is that most clusters are “deactivated” under laboratory conditions, i.e. they don’t produce anything. While we know, for example, that many streptomycetes can produce 30 or 40 different natural substances, even among the most intensively researched organisms, we do not know more than a handful of them. Helfrich and his team are applying a trick to improve their search results. “Once our algorithms have discovered interesting clusters, we optimize the latter and insert them into suitable host organisms. In most cases that reactivates the dormant cluster.”
While the team is investigating all possible bacteria, the decisive factor in this research is the type of substance the scientists are looking for. Currently, the focus is on ribosomally synthesized and post-translationally modified peptides (RiPPs). Compared to many other classes of natural products, RiPPs constitute a heterogeneous group: Many different families of RiPPs exist, most of which have nothing in common except for their basic biosynthetic principles. This makes it almost impossible to identify new RiPP families using existing genome mining tools. However, Helfrich’s lab has developed an algorithm to track them down. “The biosynthesis of RiPPs starts with the formation of a precursor peptide that is modified by certain enzymes. As a result of these changes, it is not always possible to identify the peptide as such at first glance. However, since our algorithm recognizes the special molecular fingerprint of the enzymes that modify the peptides, we are not only able to find the peptide biosynthesis gene cluster, but also new bacterial products.” The procedure can also be used for other classes of natural substances, such as terpenes. Many people recognize the characteristic odors of terpenes. The smell of summer rain, for instance, is caused by the terpene geosmin from soil bacteria. Terpenes are a promising group of natural substances: They possess excellent physical and chemical properties that otherwise only occur in synthetic materials.
Once the algorithm has found a promising cluster, the relevant natural product is isolated and examined to determine whether it really is capable of killing multidrug-resistant hospital pathogens. “We start by observing the bioactivity in an artificial nutrient medium. Afterwards we send the substance to our cooperation partners, who test it in murine infection models.” Helfrich’s team has already tracked down some natural substances that are effective against multidrug-resistant germs, as well as against fungal diseases or cancers. “Unfortunately there’s a long way to go before they reach market maturity,” Helfrich says. “That can take up to 15 years and is extremely expensive, which is why we depend on the resources provided by pharmaceutical companies that take notice of our research.”
The L-shaped proton pump
When we eat, our bodies absorb high-energy molecules, such as glucose, from our food, which are gradually broken down and ultimately used to produce adenosine triphosphate (ATP), the cell’s molecular fuel. The respiratory chain plays a major role throughout this entire process. This metabolic process takes place in the mitochondria – or to be more precise, in the inner membrane of these cell organelles. Mitochondria have an outer membrane and an inner one that is folded into cristae. Located between the two is the intermembrane space, while the matrix sits deep in the interior, bounded by the inner membrane.
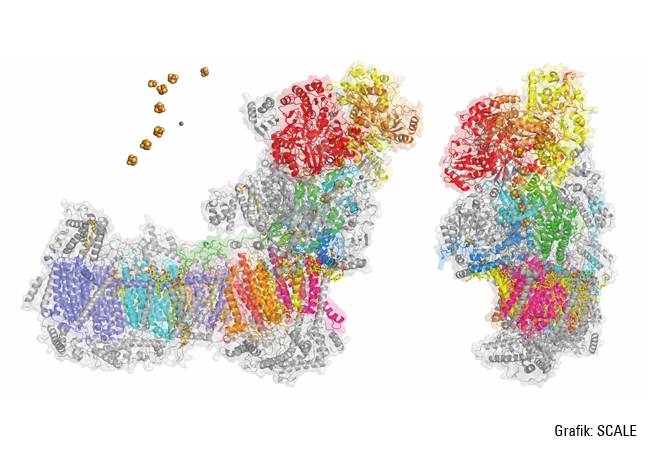
The respiratory chain consists of four protein units, the complexes I to IV. Volker Zickermann from Frankfurt University Hospital’s Institute of Biochemistry II is researching complex I, where the molecule NADH (formed by glycolysis, for example) is oxidized and releases electrons, while the molecule ubiquinone – also known as coenzyme Q10 – is reduced by picking up electrons. The energy released during this redox reaction is used to pump positively charged hydrogen nuclei, i.e. protons, through the inner mitochondrial membrane into the intermembrane space. This creates a concentration gradient, i.e. there are fewer protons inside than outside. The gradient drives the enzyme ATP synthase, part of which is made to rotate like a turbine. “So complex I is a redox-driven proton pump,” Zickermann explains. “It provides around 40 percent of the proton motive force necessary for ATP synthesis, the production of the energy-carrying molecule ATP.”
Complex I has a very characteristic shape: an L with one arm extending into the mitochondrial matrix, and another embedded in the inner mitochondrial membrane. “All redox reactions occur in the matrix arm, whereas protons are pumped in the membrane arm. This means that the two processes are physically separated, but coupled together, although it remains unclear just how the coupling works.” Zickermann wants to pinpoint exactly which mechanisms are involved in the construction and operation of this molecular machinery. To this end, he is working with the Frankfurt-based Max Planck Institute of Biophysics, using cryogenic electron microscopy – while benefiting from the vast technical progress achieved over recent years. “We can now create images of the structure of complex I at a resolution that allows us to recognize even water molecules in the membrane arm. Such resolutions enable simulations that render the dynamics in complex I visible.” In addition to suitable procedures, this also requires a suitable model organism; in this case the yeast Yarrowia lipolytica, which is deployed in several processes including the maturation of cheese. This yeast is easily genetically accessible and it has a complex I. Using Yarrowia lipolytica (among others) as the test organism, Zickermann and his team have already succeeded at clarifying some sub-steps in the biogenesis of complex I. “It has more than 40 subunits, all of which must be positioned correctly for the proton pump to work. This is the job of auxiliary proteins like NDUFAF1, which are known as assembly factors.”
Zickermann has also discovered that a protein called tafazzin is involved in the biogenesis of complex 1 in the yeast. “This was surprising, because we were familiar with tafazzin, but in a different context: It’s responsible for remodeling the lipid cardiolipin.” In eukaryotes, cardiolipin occurs almost exclusively in the mitochondria, where it is very important for the construction of the inner mitochondrial membrane, where it is believed to enable the highly convoluted shape of the cristae formed by the inner membrane. Tafazzin dysfunction in human beings causes Barth syndrome, a disease that can lead to conditions such as heart problems and muscle weakness. “The results of studies with Yarrowia lipolytica can only be applied to humans to a limited extent,” Zickermann explains. The assembly factor NDUFAF1 also occurs in our bodies, as does tafazzin, albeit not in complex I.
Zickermann would like to find out even more details about the overall process of biogenesis. Making this difficult is the fact that complex I subunits of two different genomes are coded – the genome in the cell nucleus and the mitochondrial genome – and it is not clear how the two genomes are coordinated.
Complex I is essential for energy metabolism, Zickermann says, adding that mutations can cause mitochondrial diseases like Leber hereditary optic neuropathy (LHON), which can lead to impaired vision or blindness. “Only if we fully understand the mechanisms in complex I can we also understand the pathogenesis of LHON and other diseases, i.e. why a person develops the disease.”
WHAT ELSE ARE THE SCALE SCIENTISTS PlAnNING?
In addition to the production of non-natural substances used as intelligent probes in research into the subcellular architecture, Helfrich wants to apply his expertise to investigate the subcellular compartments and the stress tolerance of the bacterial cell envelope. “Who says we can only look for natural substances? The same principles can also be applied to many other exciting questions, of which there are many at SCALE.” Volker Zickermann is also examining the processes that occur in complex I. “To be specific, we’re looking at how the biogenesis of complex I and similar complexes is connected with the structure of the membrane of cristae, i.e. the folds of the inner mitochondrial membrane” – including where and when the various substeps take place, and how the process is coordinated. https://scale-frankfurt.org
Andreas Lorenz-Meyer