Bees, wasps and ants belong to the Hymenoptera order and inject a whole cocktail of venomous ingredients when they sting. Despite their tremendous ecological and economic importance, little was previously known about the origins of their venom. Through extensive genomic studies, a team of researchers led by Dr. Björn von Reumont from Goethe University Frankfurt has now discovered that typical venomous components were already present in the earliest ancestors of Hymenoptera and must therefore have evolved before the stingers of bees and other insects. What’s more, and contrary to previous assumptions, the gene for the venom melittin is found solely in bees.

Venoms have developed in many animal groups independently of each other. One group that has many venomous species is Hymenoptera, an insect order that also includes aculeates (stinging insects) such as bees, wasps and ants. Hymenoptera is very species-rich, with over 6,000 species of bees alone. And yet, despite the great ecological and economic importance of hymenopterans, very little is known about the evolutionary development of their venoms.
By means of comparative genomics, researchers led by Dr. Björn von Reumont, who is currently a visiting scientist in the Applied Bioinformatics Working Group at the Institute for Cell Biology & Neuroscience of Goethe University Frankfurt, have now examined systematically and for the first time how the most important components of the venom of bees and other hymenopteran taxa developed in the course of evolution. The toxins are complex mixtures composed of small proteins (peptides) and a few large proteins and enzymes. Stinging insects actively inject this poisonous cocktail into their prey or attackers with the help of a special sting apparatus.
In the first step, the researchers identified which of the peptides and proteins in the venom were most prevalent in hymenopterans. To do this, they drew on information from protein databases, although this was sparse. In addition, they analyzed the proteins in the venoms of two wild bee species – the violet carpenter bee (Xylocopa violacea) and the great-banded furrow-bee (Halictus scabiosae) – as well as of the honeybee (Apis mellifera). They found the same 12 “families” of peptides and proteins in all the hymenopteran venoms analyzed. These are evidently a “common ingredient” in these venom cocktails.
In collaboration with colleagues from the Leibniz Institute for the Analysis of Biodiversity Change (LIB), the Technical University of Munich (TUM) and the LOEWE Center for Translational Biodiversity Genomics (LOEWE TBG), the research team then searched for the genes of these 12 peptide and protein families in the genome of 32 hymenopteran taxa, including sweat bees and stingless bees, but also wasps and ants such as the notorious fire ant (Solenopsis invicta). The differences in these genes, in some cases only the exchange of single letters of the genetic code, helped the scientists to determine the relationship between the genes of different species and later – with the help of artificial intelligence and machine learning – to compile a lineage of the venom genes.
The surprising result was that many of the venom genes analyzed are present in all hymenopterans. Evidently the common ancestor of allhymenopteran taxa already possessed these genes. “This makes it highly probable that hymenopterans are venomous as an entire group,” concludes von Reumont. “For other groups, such as Toxicofera, which includes snakes, anguids (lizards) and iguania, science is still debating whether the venoms can be traced back to a common ancestor or whether they evolved separately.”
Within Hymenoptera, only the stinging insects – bees, wasps and ants – have an actual stinger to administer the venom. The evolutionary old parasitic sawflies, by contrast, use their ovipositor along with their eggs to inject substances that alter their host plant’s physiology: The sirex wood wasp (Sirex noctilio), for example, not only introduces a fungus into the plant, which facilitates the colonization of the wood by its larvae, but also its own poisonous cocktail with the venom proteins examined in the study. The purpose of these proteins is to create suitable conditions in the plant for the larvae. “This means that the sirex wood wasp can also be classified as venomous,” says von Reumont.
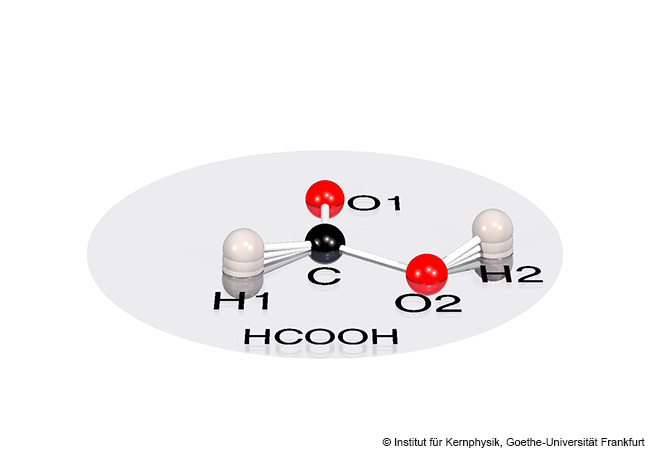
New venom components in bees are the gene for the peptide melittin and genes for representatives of the newly described protein family anthophilin-1. The fact that melittin is encoded by just one single gene came as a surprise to the researchers, as von Reumont explains: “Not only are there many different variants of melittin, but the peptide also accounts for up to 60 percent of the dry weight of bee venom. That is why science previously assumed that there must be many gene copies. We were able to disprove this quite clearly.” Because they found the melittin gene only in bees, the researchers also invalidated the hypothesis that it belongs to a group of venom genes postulated for stinging insects called aculeatoxins. Von Reumont is convinced: “This shows us once again that genome data are the only way to draw meaningful conclusions about the evolution of venom genes.”
The Frankfurt study is the first one to show for an entire insect group with around one million species where venom genes originated and how they have developed. It provides a starting point for tracing the evolution of venom genes in the ancestors of Hymenoptera as well as specializations within the group. However, to be able to perform comparative genomics on a large scale, analysis methods for the partly very large protein families must first be automated.
Publication: Ivan Koludarov, Mariana Velasque, Tobias Senoner, Thomas Timm, Carola Greve, Alexander Ben Hamadou, Deepak Kumar Gupta, Günter Lochnit, Michael Heinzinger, Andreas Vilcinskas, Rosalyn Gloag, Brock A. Harpur, Lars Podsiadlowski, Burkhard Rost, Timothy N. W. Jackson, Sebastien Dutertre, Eckart Stolle, Björn M. von Reumont: Prevalent bee venom genes evolved before the aculeate stinger and eusociality. BMC Biology, (2023) doi.org/10.1186/s12915-023-01656-5