A team around Ivan Dikic and Mike Heilemann discovers an inflammatory signaling platform which may lead to novel antibiotic therapies
With drug resistance being on the rise worldwide, bacterial infections pose one of the greatest global threats to human health. By deciphering the host-pathogen interaction on a molecular level, researchers hope to pave the way for new therapies. Studying the cell’s reaction to Salmonella, scientists from Goethe University Frankfurt have now made a critical discovery to this respect.
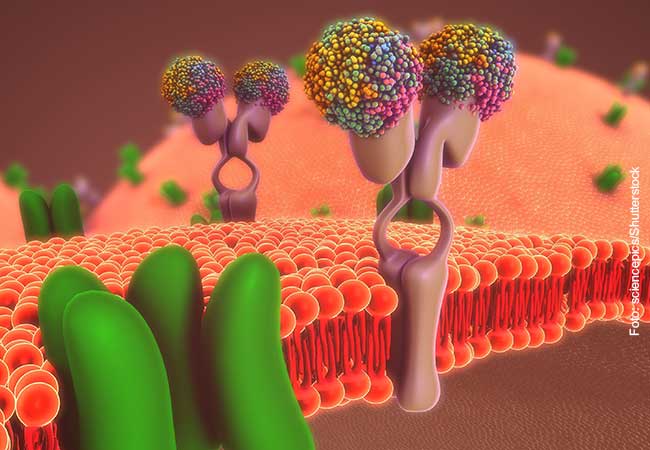
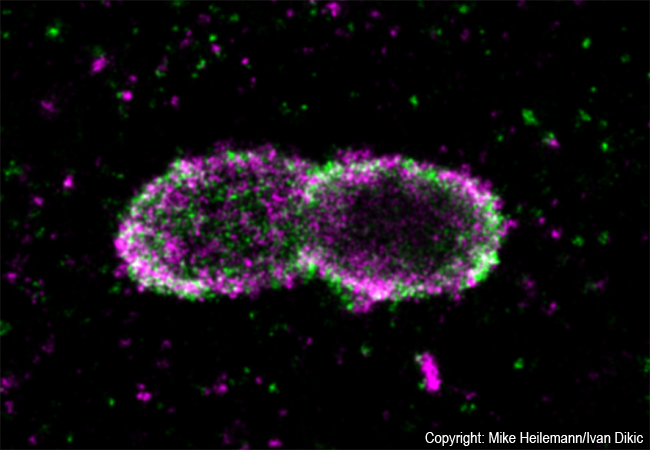
All bacteria have developed clever mechanisms for survival and propagation within host cells. Salmonella are a typical example: usually they hide in membrane-bound particles with only very few bacteria escaping to the cell’s interior. Those escapees are extremely dangerous as they proliferate and spread at enormous speed. To stop such an invasion, cells have developed very effective defense strategies. An interdisciplinary team around Prof. Ivan Dikic (Institute of Biochemistry II) and Prof. Mike Heilemann (Institute of Physical and Theoretical Chemistry), both from Goethe University Frankfurt, now studied such a cellular defense mechanism by visualizing protein patterns at the near-molecular level.
Protein chains relay pro-inflammatory signals
Upon bacterial invasion, cells react fast: They flag escaped bacteria with a small protein called ubiquitin, which is known to regulate numerous cellular processes. The attached flags contain chains of differently linked ubiquitin molecules, resulting in a secret code, which has so far only partially been decoded. Similar to mobile transmission towers, these ubiquitin chains relay specific signals from the surface of the bacteria into the cell.
Employing super-resolution microscopy, the Frankfurt team now succeeded with visualizing different ubiquitin chains on the bacterial surface and analyzing their molecular organization in detail. They discovered that one chain type, so called linear chains, plays an essential role during a bacterial invasion. Linear ubiquitin chains trigger degradation of bacteria and kick off an inflammatory signaling cascade which results in restricting bacterial proliferation. In addition, the researchers identified the enzyme Otulin as an important regulator capable of limiting this reaction – a very important notion considering the fact that excessive inflammation is one of the major causes of tissue damage following bacterial infection.
Signaling the cells’ need for pathogen defense is just one important role of ubiquitin. The small protein is also involved in development and progression of inflammatory and neurodegenerative diseases as well as of cancer. Until now, however, very little is known about how small errors in the ubiquitin system contribute to these serious human diseases, and how the system can be targeted pharmaceutically.
These new findings pave the way for many follow-up projects which may ultimately lead to novel therapeutic approaches. Very recently, Ivan Dikic obtained one of the prestigious ERC Advanced Grants of 2.5 M € in which he will investigate the role of ubiquitin in modulating the host-pathogen interaction in more detail.
The work of the Frankfurt team is an excellent example for interdisciplinary collaboration and was enabled by funding of several large research networks, e.g. the Cluster of Excellence Macromolecular Complexes, the CRC 1177 on selective autophagy and the LOEWE ubiquitin network. The results are now published in the latest online issue of Nature Microbiology, back-to-back with complementary insights generated by colleagues in Cambridge (UK).
Publication: van Wijk SJ, Fricke F, Herhaus L, Gupta J, Hötte K, Pampaloni F, Grumati P, Kaulich M, Sou Y, Komatsu M, Greten F, Fulda S, Heilemann M, Dikic I. Linear ubiquitination of cytosolic Salmonella Typhimurium activates NF-κB and restricts bacterial proliferation. Nature Microbiology 2017, doi 10.1038/nmicrobiol.2017.66.
Source: Press Release 19/05/17