The respiratory chain plays a central role in energy metabolism of the cell. It is localized in mitochondria, the cell´s own power plants. In a new study, researchers from Goethe University, the Max Planck Institute of Biophysics and the University of Helsinki have determined the high-resolution structure of a central component of the respiratory chain, mitochondrial complex I, and simulated its dynamics on the computer. These findings both support basic research and enhance our understanding of certain neuromuscular and neurodegenerative diseases that are linked with mitochondrial dysfunction.
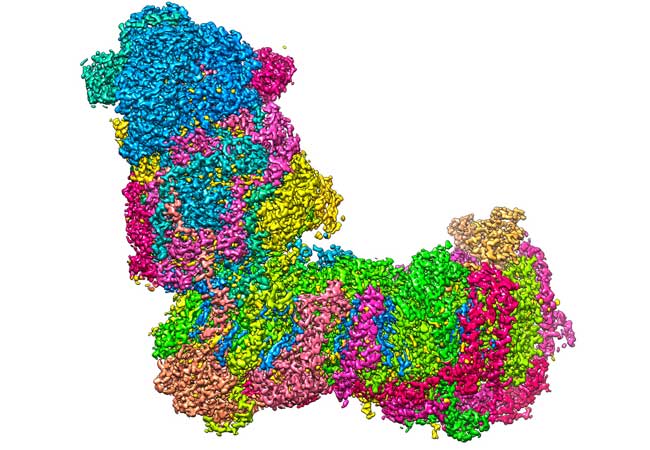
A bit like a boot: The L-shaped structure of mitochondrial complex I at a resolution of 2.1 Ångström (0.00000021 millimetres), captured with a cryo-electron microscope.Image: Janet Vonck, MPI of Biophysics
All vital processes require a constant supply of energy. In the cell, the chemically “charged” molecule ATP is the main provider of this energy. The ATP power packs are produced, among others, in specialised small organs (“organelles”) of the cell, the mitochondria.
There, the protein complexes of the respiratory chain pump hydrogen ions (protons with a positive charge) from one side of the inner mitochondrial membrane to the other (“uphill”), creating a chemical concentration gradient and an electrical voltage. The protons “flow downhill” along this electrochemical gradient through a kind of turbine that generates useful energy for the cell in the form of ATP.
One of the proton pumps in the first step of the process is a large, L-shaped biomolecule, mitochondrial complex I (in short: complex I). Its horizontal arm is anchored in the membrane. The vertical arm binds the electron carrier molecule NADH, which is produced during metabolic breakdown of sugar and other nutrients. Complex I catalyses the transfer of electrons from NADH to ubiquinone (Q10), and the energy released in this reaction is used to drive the proton pump.
The research team from Goethe University and the Max Planck Institute of Biophysics in Frankfurt used cryo-electron microscopy to determine the 3D structure of complex I at high resolution. The researchers were able to show that water molecules in the protein structure play an important role for establishing proton translocation pathways.
The high-resolution structural data enabled colleagues at the University of Helsinki to conduct extensive computer simulations, which show the dynamics of the protein structure during its catalytic cycle.
Dr Janet Vonck from the Max Planck Institute of Biophysics explains: “Our study delivers new insights into how a molecular machine in biological energy conversion works.” Professor Volker Zickermann from the Institute of Biochemistry II at Goethe University says: “This knowledge can contribute to a better understanding of certain mitochondrial diseases, such as loss of vision in Leber hereditary optic neuropathy.”
Publication: Kristian Parey, Jonathan Lasham, Deryck J. Mills, Amina Djurabekova, Outi Haapanen, Etienne Galemou Yoga, Hao Xie, Werner Kühlbrandt, Vivek Sharma, Janet Vonck, Volker Zickermann: High-resolution structure and dynamics of mitochondrial complex I – Insights into the proton pumping mechanism. Sci Adv. 2021 Nov 12;7 (46) https://www.science.org/doi/10.1126/sciadv.abj3221